MicroRNAs |
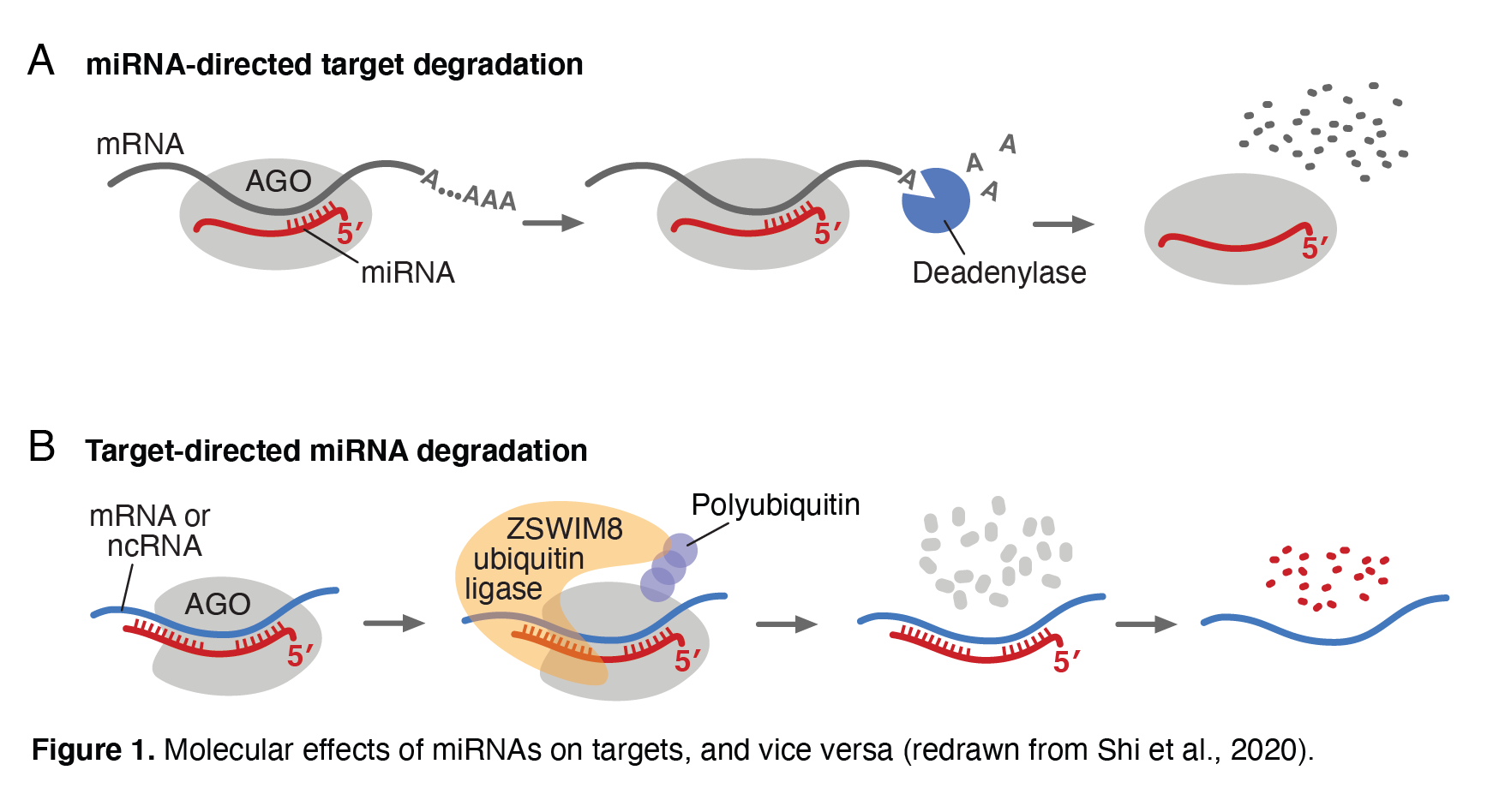
MicroRNAs are short regulatory RNAs that direct widespread gene repression in the cells of humans and other animals. Our lab has helped to define microRNAs, their regulatory targets, and their molecular and regulatory consequences in animals and plants. We continue to study these regulatory RNAs, as exemplified below. Each microRNA associates with an Argonaute (AGO) protein to form a silencing complex in which the microRNA pairs to sites within target mRNAs, and AGO recruits machinery that represses the targeted transcript (Figure 1A). We have recently acquired high-throughput biochemical measurements to evaluate the affinities between purified AGO–microRNA complexes and hundreds-of-thousands of RNA-sequence possibilities. These results are revealing differences between the targeting preferences of different microRNAs, which is helping us better predict which mRNAs are most affected by each microRNA (predictions available at targetscan.org). We have also been exploring how microRNAs themselves are regulated. Other labs have shown that pairing between a microRNA and a target with unusually extensive complementarity can trigger decay of the microRNA, inverting the typical regulatory logic. This phenomenon, termed target-directed microRNA degradation (TDMD), is exploited by some viruses, which express transcripts that direct the degradation of host microRNAs that would otherwise impede viral replication. We and others discovered that the ZSWIM8 E3 ubiquitin ligase is required for TDMD (Figure 1B), and we have gone on to show that this ubiquitin ligase is used by cells of mammals, insects, and nematodes to shape the levels of many endogenous microRNAs.mRNAs
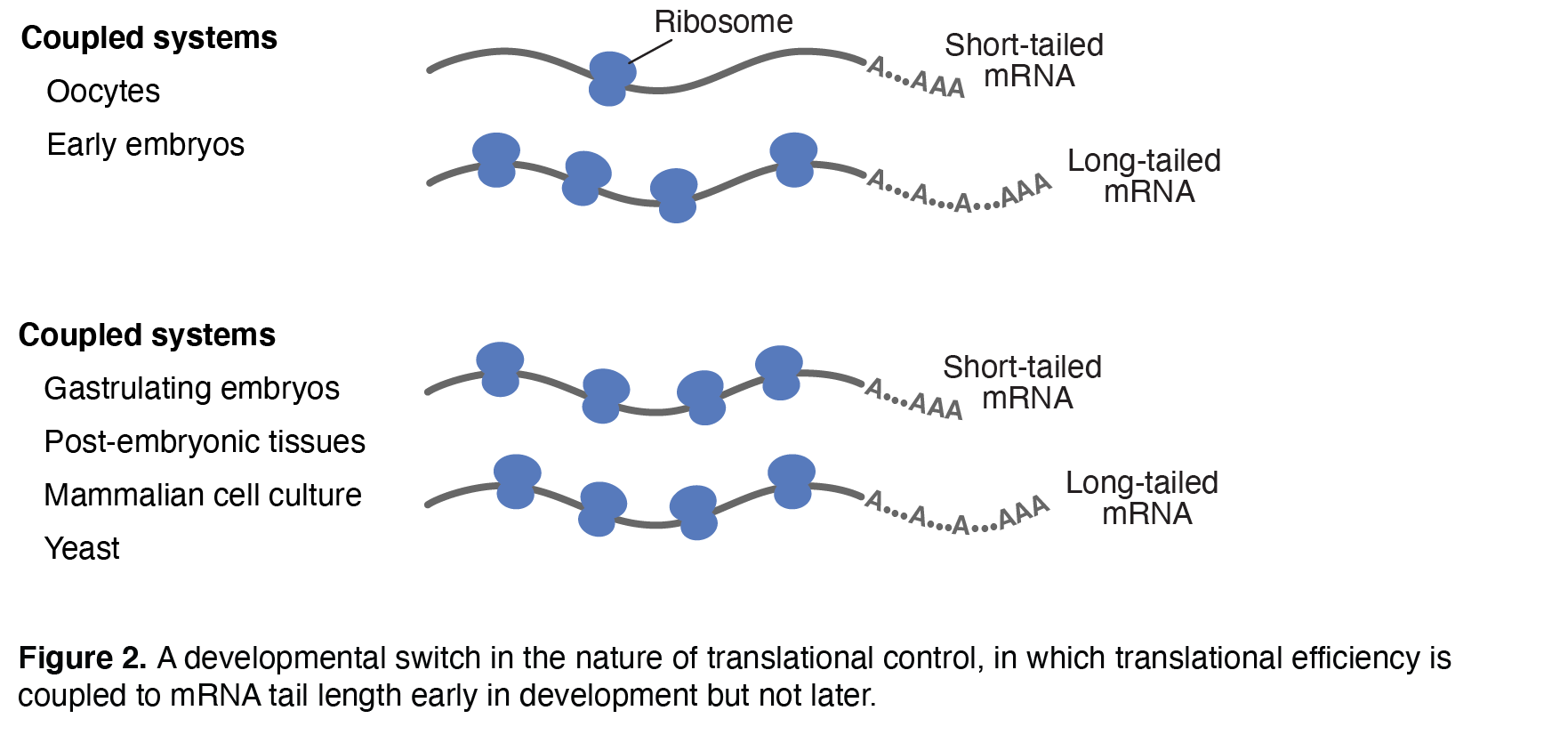
Our interest in microRNAs has expanded to the mRNAs that they regulate, particularly their untranslated regions (UTRs) and poly(A) tails, and the roles that these regions play in recruiting/mediating post-transcriptional regulatory phenomena. For example, we developed high-throughput methods for measuring the lengths of poly(A) tails appended to the ends of most eukaryotic mRNAs. Using these measurements, we found that mRNA tail length and translational efficiency are strongly coupled in early fish, frog and fly embryos but that this coupling diminishes later in development and is absent in non-embryonic samples—revealing a previously unrecognized developmental switch in the nature of translational control (Figure 2). We have recently determined the molecular underpinnings of this developmental switch, and we have applied our methods for measuring poly(A)-tail lengths to discover principles of mammalian mRNA metabolism. Other projects have identified structured regions of mRNAs and investigated their potential functions. For example, we identified thousands of mRNA regions that can fold into G-quadruplexes and then measured their folding state in cells, which revealed that RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. In addition, we found more conventional folding of mRNA 3′-end regions in human cells and showed that these structures enhance mRNA processing and stability. We also discovered that under certain stressed conditions, some yeast introns accumulate as excised linear molecules that help to regulate growth.Current and Future ResearchWe are building on these and other discoveries to enhance the molecular and biological understanding of microRNA targeting, target-directed microRNA degradation, long noncoding RNAs, differential metabolism of cellular and viral mRNAs, translational control, and other aspects of post-transcriptional gene regulation. | ||

About - The Bartel Lab of Whitehead Institute For Biomedical Research
Last update: Nov-29-2021
Bartel laboratory - © 2009 Whitehead Institute for Biomedical Research
9 Cambridge Center Cambridge, MA 02142 USA
Web admin: jgulling{at}mit[dot]edu
Accessibility