mirscanExecute.py performs the function of applying scores to each in a set of candidate hairpins according to a pre-defined scoring matrix. The evaluation of hairpins is done using a set of criteria implemented in some criteria-formatted file (referred to here as mirscanCriteria.py). The same mirscanCriteria.py file should also have been used for the training step, performed by mirscanTrainer.py, that generated the scoring matrix. The feature evaluation functions of mirscanCriteria.py implement an author's ideas regarding what is important about a miRNA hairpin (base pairing, conservation, etc.). Such ideas are expected to differ between different authors, and even to change for a single author depending on time and context. The design of this system allows a programmer to implement a given set of ideas only once, in the context of mirscanCriteria.py. The inheritance of that implementation by both mirscanTrainer.py and mirscanExecute.py facilitates the beginning-to-end process of mirscan-based candidate evaluation.
An example of this flexibility is provided in the current study by Ruby et al. (Genome Res, 2007) in the change from considering a candidate as a single sequence to a candidate as a pair of (presumably orthologous) sequences. The two concepts of a candidate were each implemented as a separate instance of the mirscanCriteria.py script (oneSeq.RubyEtAl.py for single sequence candidates, twoSeq.RubyEtAl.py for sequence pair candidates). On this site, the user is provided with both of those modules, as well as prototype versions of each (oneSeq.prototype.py and twoSeq.prototype.py) to which feature objects can be added as desired.
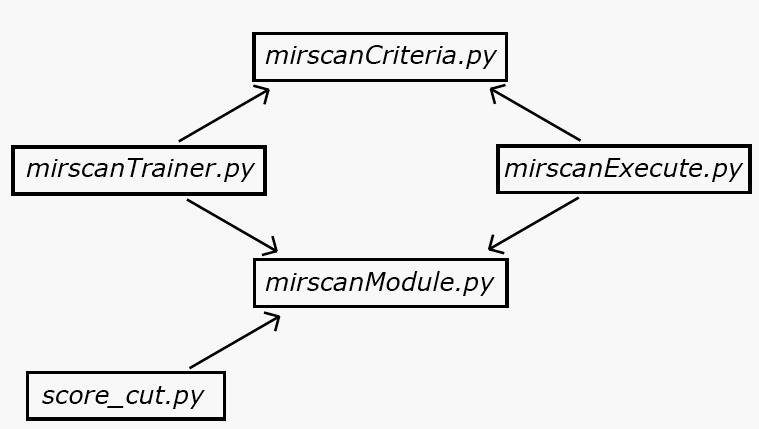
Above are the fundamental dependencies of the scripts comprising the mirscan system. All scripts depend on mirscanModule.py, where most of the core functionality used across the system is implemented. mirscanTrainer.py and mirscanExecute.py additionally depend on the mirscanCriteria.py files that are used to evaluate miRNA hairpin candidates. These two dependencies are what gives the mirscan system its flexibility. In modifying or expanding mirscan's functionality, the user may create additional dependencies. For instance, the function get_folds in the provided mirscanModule.py depends on the RNAfold application from the Vienna RNA Package being installed on the host computer system. This dependency could be removed or changed if the implementation of get_folds were changed. The most independent component of the system is score_cut.py, which only depends on mirscanModule.py for the file-parsing functions that it uses. A more complete listing of the dependencies for each module is provided below: